Difference between revisions of "POC Conf. Call 9-10-13"
| Line 30: | Line 30: | ||
*''' [https://sourceforge.net/p/obo/plant-ontology-po-term-requests/555/ vascular leaf development stage (new PO:0025570)]''': A leaf development stage (PO:0001050) that begins with the onset of vascular leaf initiation stage (PO:0007040) and ends with the termination of the vascular leaf senescence stage (PO:0001054). | *''' [https://sourceforge.net/p/obo/plant-ontology-po-term-requests/555/ vascular leaf development stage (new PO:0025570)]''': A leaf development stage (PO:0001050) that begins with the onset of vascular leaf initiation stage (PO:0007040) and ends with the termination of the vascular leaf senescence stage (PO:0001054). | ||
| − | ''Add comment on how every stage can end in accidental death'' | + | ''Add comment on how every stage can end in accidental death. Also mention something death being the conical end point'' |
*non-vascular leaf development stage (new PO:id): A leaf development stage (PO:0001050) that begins with the onset of non-vascular leaf initiation stage and ends with the termination of the non-vascular leaf senescence stage. | *non-vascular leaf development stage (new PO:id): A leaf development stage (PO:0001050) that begins with the onset of non-vascular leaf initiation stage and ends with the termination of the non-vascular leaf senescence stage. | ||
Revision as of 08:47, 11 September 2013
POC meeting, Skype Conference Call; Date: Tuesday Sept 10th, 2013 10am PST/1pm EST
In attendance: Laurel Cooper (OSU), Justin Elser (OSU), Justin Preece (OSU), Brian Atkinson (OSU), Dennis W Stevenson (NYBG), Pankaj Jaiswal (OSU), Barry Smith, Marie Alejandra Gandolfo (Cornell)
Absent: none
- please see the Items_for_future_meetings page, will be moved here before the meeting
multi-tissue plant structure development stage (PO:new)
- Needs a parent term: multi-tissue plant structure development stage (PO:new), then the definitions can be re-written, currently a direct child of plant structure development stage (PO:0009012). This would also be the parent for seed development and plant organ development (see below)
Accepted, will post proposed definition on SF tracker
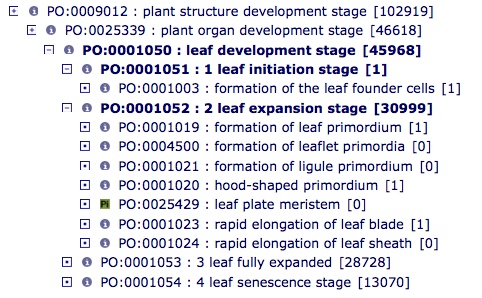
leaf development stage (PO:0001050)
- See Google Doc for revisions in progress.
- Need to look into non-vascular leaf development and see if it applies there as well, or if we will need separate terms for vascular and non-vascular leaves.
- leaf development stage (PO:0001050): A phyllome development stage (PO:0025579) that has as a primary participant a leaf (PO:0025034).
Current structure: All these will be renamed "vascular leaf...."
- Links to Relevant References
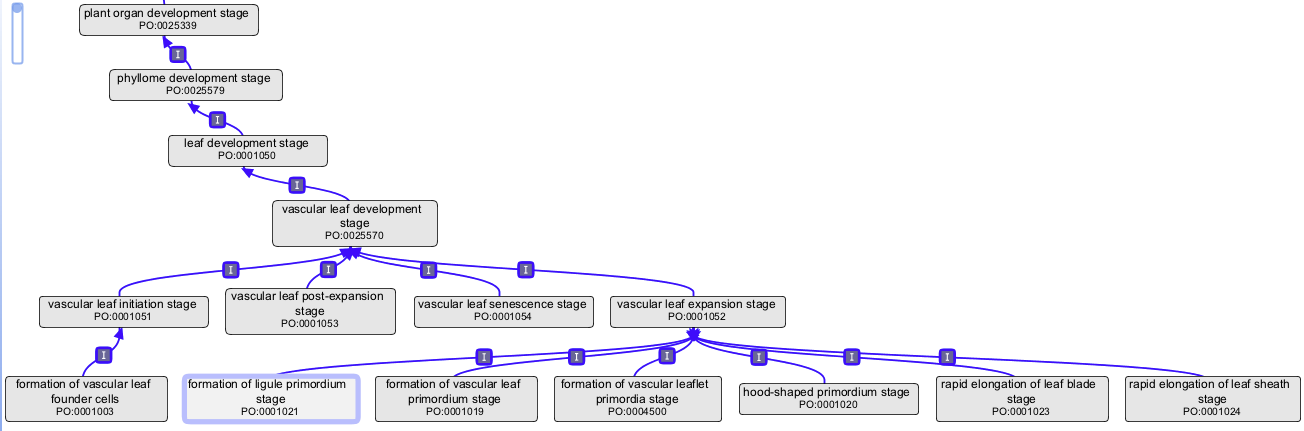
vascular leaf development stage and child terms
Proposed new term and definition:
- vascular leaf development stage (new PO:0025570): A leaf development stage (PO:0001050) that begins with the onset of vascular leaf initiation stage (PO:0007040) and ends with the termination of the vascular leaf senescence stage (PO:0001054).
Add comment on how every stage can end in accidental death. Also mention something death being the conical end point
- non-vascular leaf development stage (new PO:id): A leaf development stage (PO:0001050) that begins with the onset of non-vascular leaf initiation stage and ends with the termination of the non-vascular leaf senescence stage.
- Need to do more research on the development of the non-vascular leaf
see also: moss phyllides, moss info and Background_info_on_Physco_biology_and_culture.
New/revised child terms under vascular leaf development stage (PO:0025570)
- vascular leaf initiation stage (PO:0007040): A vascular leaf development stage (PO:0025570) when cells of the peripheral zone (PO:0000225) in the shoot apical meristem (PO:0020148) transition from indeterminate growth to determinate growth and ends at the beginning of the vascular leaf expansion stage (PO:0001052).
Comment: Cells in the primordium are not yet differentiated.
- vascular leaf expansion stage (PO:0001052): A vascular leaf development stage (PO:0025570) that begins with cell differentiation and cell expansion forming the leaf lamina (PO:0020039) and ends with the onset of the leaf fully expanded stage (PO:0001053).
- new comment: The majority of leaf expansion is due to cell enlargement. The leaves of some plants may not have a conspicuous midrib or rachis.
- vascular leaf post-expansion stage (PO:0001053): A vascular leaf development stage (PO:0025570) that begins when the vascular leaf (PO:0009025) is fully expanded and cell expansion has ceased and ends with the onset of the vascular leaf senescence stage (PO:0001054).
- vascular leaf senescence stage (PO:0001054): A vascular leaf development stage (PO:0025570) that begins with the formation of a abscission zone at the base of a vascular leaf (PO:0009025) and ends with leaf separation and death.
Note: Will have the new preceded_by relation added to order them
Children of vascular leaf initiation stage (PO:0007040)
- formation of vascular leaf founder cells (PO:0001003): A vascular leaf initiation stage (PO:0007040) when the cells of the shoot apical meristem (PO:0020148) form a vascular leaf anlagen (PO:0025431).
Comment: The leaf primordium is not yet distinguishable.
Children of vascular leaf expansion stage (PO:0001052)- revised names and definitions:
- formation of vascular leaf primordium stage (PO:0001019): A vascular leaf expansion stage (PO:0001052) when the shoot apical meristem (PO:0020148) forms a vascular leaf primordium (PO:0000017) on the peripheral zone (PO:0000225).
- formation of vascular leaflet primordia stage (PO:0004500): A vascular leaf expansion stage (PO:0001052) at which a leaflet primordium (PO:0025481) is initiated on the leaf rachis (PO:0020055) of a compound leaf (PO:0020043).
- formation of ligule primordium stage (PO:0001021): A vascular leaf expansion stage (PO:0001052) when conical P3 leaf completely encloses SAM, and the ligule (PO:0020105) primordium is visible.
- hood-shaped primordium stage (PO:0001020: A vascular leaf expansion stage (PO:0001052) when a hood-shaped primordium is formed, partially enclosing P1 and SAM, as in Poaceae.
- rapid elongation of vascular leaf lamina stage (PO:0001023): A vascular leaf expansion stage (PO:0001052) during which a rapid elongation of the leaf lamina (PO:0020039) occurs.
- elongation of leaf sheath stage (PO:0001024): A vascular leaf expansion stage (PO:0001052) during which a rapid elongation of the leaf sheath (PO:0020104) occurs.
OBO Foundry Review of Plant Ontology
Principle 1: The Ontology is Open
Some improvement desirable
"The self-review states that the PO is released under a Creative Commons license. However, it is not specified which Creative Commons license has been chosen. The ontology file does not contain a statement about the license or the policy for reuse of the ontology. However, the information is available on the website e.g. at http://plantontology.org/docs/otherdocs/principles_rationales.html: Plant Ontology by Plantontology.org is licensed under a Creative Commons Attribution-NoDerivs 3.0 United States License."
"We recommend that the exact license chosen be included in an additional ontology annotation in the ontology file."
"According to the OBO Foundry principle 1 documentation, this should make use of two annotations using the following properties:
- dc:license
- rdfs:comment
The license type goes in the dc:license field. The mode of attribution goes in the rdfs:comment field. "
"While 'Creative Commons 3.0 CC-by license' is recommended, this is not mandatory and therefore the chosen attribution-noderivs license is also compatible with the Open principle. However, we urge you to consider weakening the license to one that encourages sharing and reuse without constraint to further the adoption by the community and the value of the resource for the community."
Principle 2: Common Format
Pass
The ontology is available in both OBO and OWL formats. It should be noted that the OBO form of the ontology contains semantics which are not yet available in the OWL form (i.e. the treat-xrefs-as-isa and treat-xrefs-as-genus-differentiae constructs). These are supported by the Oort release manager and can be instantiated in OWL versions of the ontology. See https://code.google.com/p/owltools/wiki/OortOptions
Principle 5: Clearly Delineated Content
Some improvement desirable
"There are several sub-issues that are contained in this principle.
Firstly, the ontology needs to have a clearly delineated scope as expressed in a statement of scope. PO does have a clearly delineated scope, and this scope does not substantially duplicate the scope of any other OBO Library ontology.
Secondly, the ontology needs to have a clear upper level with root terms having definitions that indicate the content of the sub-ontologies included in the ontology. Here, we have some concerns as detailed below.
Thirdly, the ontology should not contain terminology that overlaps with other OBO ontologies except where content has been imported for reuse in specific contexts appropriate to the domain of the ontology.
Finally, the ontology should provide adequate coverage with respect to its stated domain so as to justify its use in applications. Concerns about the upper levels of the plant ontology:
"The definition of the root term ‘plant anatomical entity’ appears intended to be a plant specific subclass of the CARO term ‘anatomical entity’ and is defined as “An anatomical entity that is or was part of a plant.” The CARO class states that an anatomical entity is part of or located in an organism (or virus or viroid in the upcoming release of CARO), however here you include things that were once part of a plant. In OBI or ERO, such entities would be considered specimens or samples. Can you explain why this is needed? If it is justified, there is some cross-ontology coordination that should be performed.
"There is a need to define the high-level distinction between plant structure vs. portion of plant substance better: currently ‘plant structure’ is defined as anatomical entity that is or was part of a plant; but substances are also (or were also) part of a plant. This is likely because these are intended to be subclasses of the CARO classes, that have more specific differentia, however, there is no mireot or subclass assertion to these classes other than in the comment field. Explicit use of the CARO classes will help differentiate these top-level nodes, and inclusion of their logical class definitions can help with error-checking."
" If you consider the above point on the definition of ‘anatomical entity’ referring to in vivo entities, you could consider moving ‘in vitro plant structure’ to be its own root outside anatomical entity, as it would technically be a derived structure and not part of or located within an organism (or are most of these simple plants cultured in vitro?). Efforts have been made recently in the CL and OBI to represent such things (e.g. “in environment” vs. “ex environment”, you could potentially leverage some of these design patterns and/or coordinate with them. Other concerns from somewhat random sampling:
"‘cultured plant embryo’ has two asserted superclasses. While asserted multiple inheritance is debatable in terms of ontology maintenance or ontological rigor (GO has a lot of this), in this case the example ensues because of a lack of logical class expression on this class. For example, this class could instead leverage a class restriction that states ‘plant embryo’ and is_specified_output of some in vitro plant culturing. In this way, you would infer the second parent, ‘in vitro plant structure’ (dependent on how you define things as per above). This would mean higher quality axioms, easier ontology maintenance, and interoperability with other ontologies such as OBI."
Principle 6: Textual Definitions
- Some improvement desirable
We applaud the fact that the ontology does contain text definitions for all terms included in the ontology (see Appendix: Metrics) and for its extensive synonyms in numerous languages. However, we note that some of the text definitions do not exactly match the formal definitions as given in equivalence class or subclass axioms (specific examples contained below) and furthermore that not all of the textual definitions are as clear as they could be e.g. by following the genus differentiae pattern (examples below). We advise continued evolution of the textual definitions contained in the ontology towards greater clarity for the benefit of all users, and gradual inclusion of a greater number of equivalent class definitions. Specific points:
- Definitions such as that for ‘microgametophyte’, referencing IDs within the text, should be converted into logical definitions.
- ‘pollen’ is textually defined as located_in some ‘pollen sac’ and logically it has the relationship part_of some pollen sac. This is an inconsistency with respect to the normal usage of these two relationships.
Principle 7: Use of Relations
- Some improvement desirable
For the most part, the PO uses RO relations. The relation that is PO specific is provided with adequate documentation in the form of a definition and comment. Some relations are, however, defined in the PO that are within RO or BFO scope and yet do not have appropriate cross-references/Mireoted URIs. These include ‘has_participant’, ‘located_in’ and ‘participates_in’. This should be addressed, and full inclusion of such properties will facilitate the use of their logical properties for error checking.
Specific points:
- derives_by_manipulation_from is described as a subproperty of derives_from, whereby there has been a human processing step. This appears to be some kind of shortcut but isn’t yet logically defined as such. We would urge PO to work with OBI to leverage the work they have done in defining organismally derived entities, whereby such entities are the outcome of a ‘material processing’ process, has_specified_output the processed entity, and are derived_from the parent anatomical entity (for example see ‘urine specimen’).
- Plant developmental stage classes do not yet have precedes relations for temporal ordering. Implemented in Version #20
- Consider using the stage relations (for example, existence_starts_during) from Uberon and similar to the zebrafish anatomy ontology, to relate organism developmental stages to the organismal entities at those stages. For example, ‘plant embryo’ could be defined as ‘whole plant' and (existence_ends_with some 'embryo stage') and (existence_starts_with some 'embryo stage') rather than use of the participates relation.
Principle 8: Documentation
- Pass
- Extrinsic: The ontology is well documented with an extensive website and several publications.
- Intrinsic: The ontology has high quality text definitions for every term.
- Specific points:
- There is some conflation in the use of annotation properties. A lot of information is placed in the comments field, including examples of usage, subclass assertions, and stage or taxon specific considerations. You may wish to break these out into more specific annotation properties. See IAO-core metadata and/or Uberon for some options, and please feedback to IAO if you desire a standard annotation property that isn’t yet available.
- In some cases, definitions are attributed to “POC:curators”. We’d urge you to consider use of specific persons or sources for attribution, and as the Foundry moves towards the use of ORCID IDs, to include these as part of your attribution strategy.
- While we applaud the reference of tracker items directly in the ontology, these items can be included as URLs such that the link is easily made directly from the ontology, for example see ‘leptome’ to “OBO_SF_PO:3295055”.
- Similarly with the PO_REFs- please make these URLs or imports so that they can be easily located. It is likely that a new standardized annotation property for this purpose should be created and used across the Foundry.
NSF PI meeting Sept 4th-6th in DC
- PJ and DWS attended
Any news?
Upcoming meetings and Presentations 2013-2014:
- The Plant Genomics Congress USA, St Louis, September 23rd & 24th, 2013
No one from PO is planning to attend
- PAG 2014 January 11-15, 2014 - San Diego, CA, USA
- PJ has been contacted by PAG organizers; Suggest slightly new focus "Plant Ontologies and Systems Biology"
- Ideas for speakers- someone from GO and others?>>
- Registration is now open: PAG 2014. Early reg'n till Nov. 1st
Next meeting scheduled for Tuesday, Sept 24th, 2013 at 10am Pacific/1pm Eastern
-MAG has requested the meeting be moved to 10:30am Pacific/1:30 pm Eastern- would that works for the others in the group?