POC Technical Issues Page
This page provides a record of the technical issues and software needs of the POC. When requesting new software, please provide an example of why the software is needed. Also provide the date that the request was first made. If the problem has been solved (either by creation of new software, or through some other means, please provide information on the solution here.
Problem: Loading has_part relations in the Plant Ontology File
Original request: This became a problem with with the March 2011 release. (Actually, this was a problem when we did the Jan 2011 release, as the has_part relation had to be stripped out).
The Problem:
-The has_part relation causes a 'circularity or cycle' when loading the Ontology file into the database (not onto the browser as we originally thought?).
-Note: Our collaborators (eg; SGN), who uses a Chado schema, also have to remove them (LC).
-What exactly is causing the problem with loading? Is it actually the has_part relations themselves? (note: removing them allows the file to be loaded with no problems)
-JP: Why does the AmiGO data loading process choke on circular has_part::part_of relations when OBO-Edit doesn't? Does AmiGO's data-loading script perform a reasoning validation?
Questions:
- Do the problems arise when we have reciprocal part_of and has_part relations (or reciprocal part_of and develops_from relations) ?
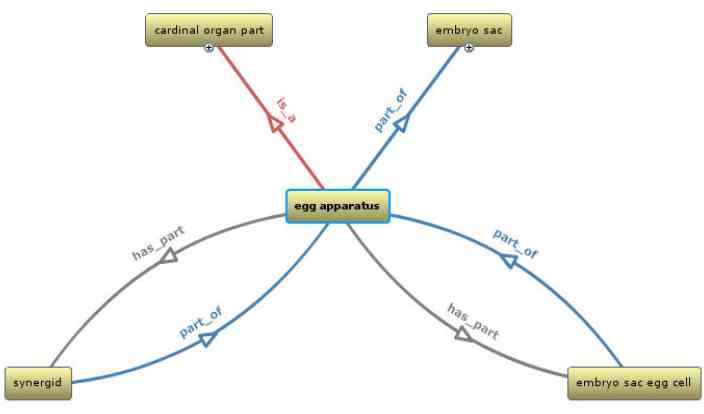
For example, embryo sac egg cell is part_of egg apparatus and egg apparatus has_part embryo sac egg cell.
- Uberon has a similar situation with mammary gland secretes milk and milk secreted_by mammary gland. Does that cause a problem?
- Also, PO has reciprocal disjoint_from relations, and they don't seem to cause a problem,
Example- why it is needed:
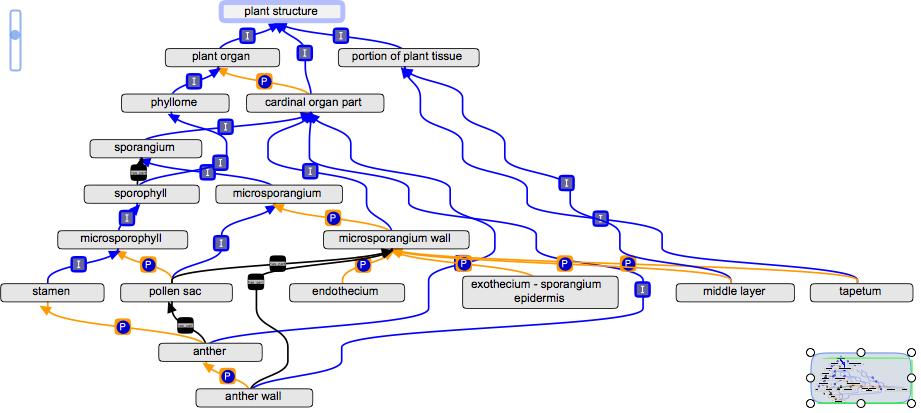
Below is an example of how the has_part relation would be used in the PO to deal with the parts of the sporangium wall:
The sporangium wall is basically the same throughout vascular plants, so this proposal creates a new general term "microsporangium wall". The part_of children are part of microsporangium wall (sort of like the part_of children of leaf). Anther wall has_part sporangium wall, since an anther is composed of multiple sporangia. In the current version of the PO, parts of the microsporangium wall are parts of the anther wall, but that only works for angiosperms.
Note: With this approach, we not only need the software to be able to handle the relations, we need a way to assure that annotations move through has_part relations
Possible Solutions:
3-16-11 test RW: Removed the has_part relations from the reciprocal has_part and part_of relations from about 15 terms in the plant_anatomy.obo file.
JE loaded the file into the database and the onto the AmiGO browser with no problems has_parts in the PO structure ontology
3-17-11 RW removed the following relations from the developers' version of plant_ontology.obo and put it on SVN.
Removed has_part (left in reciprocal part_of) relations from:
Abscission zone > protective layer, separation layer
Basal endosperm transfer layer > basal endosperm transfer cell
Egg apparatus > embryo sac egg cell, synergid
Leaf stomatal complex > leaf guard cell, leaf stomatal pore
Phyllome stomatal complex > phyllome guard cell, phyllome stomatal pore
Shoot apex > shoot apical meristem, leaf primordium
Stomatal complex > guard cell, stomatal pore
Removed develops_from relations from (left in reciprocal has_part):
Megaspore > tetrad of megaspores
Microspore > tetrad of microspores
Note: We will need to will check into it which way would have been better biologically.
The following has_part relations are still in the file:
collective organ part structure > cardinal organ part
collective plant structure > plant organ
coma > seed trichome
embryo axis > hypocotyl, root meristem
first order inflorescence axis > peduncle
inflorescence > flower
infructescence > fruit
meristem > meristematic cell
second order inflorescence > flower, second order inflorescence axis
second order infructescence > fruit, second order infructescence axis
tetrad of megaspores > megaspre
tetrad of microspores > microspore
theca > pollen sac
We can try to load this file onto the dev or beta browsers, to see if it works.
3-17-11 test:
Still need to determine if this fix works for Chado users.
Alternative solutions
!. Have all of the parts of a structure be only part of the most general class, then write a script so annotations to any part_of child of that class also go to the correct specific is_a child (as describe above for leaf). This would allow us to use the current software such as Amigo and Chado, but would require additional software and possibly special coding in the obo file for all terms that need special treatment.
2. Have specific part of children for all of the specific is_a children. This would not require any changes to the existing software or new scripts. However, it would quickly lead to term inflation. Also, it would not be biologically accurate, since the is_a children would be based largely on the taxon in which they occur, not on real differences in the structure.
3. Use the PO as an uber-ontology for plants, then create taxon-specific sub-ontologies that cross-reference the main ontology. This would create a similar number of terms as solution number 2, but they would not all be in one ontology, so it may be more manageable. Would create problems in syncing the ontologies, and could make cross-species comparisons more difficult. Of course, even within a single taxon (like angiosperms) there may be a need to use the has_part relation (like inflorescence has_part flower).
Need: Script or an Algorithm for Assigning Annotations
Original request: March 1, 2011
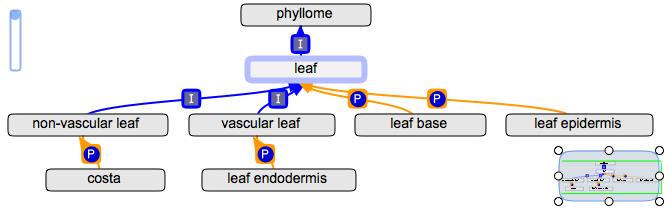
Example: For gathering annotations from parts of leaf to either vascular leaf or non-vascular leaf
Why this is needed:
At the POC_Conf._Call_3-1-11 we agreed that in general, it is better to have the part_of children of leaf only be part of the general term leaf, rather than create new terms for vascular parts of and non-vascular parts of. Terms that only occur in one or the other (like leaf endodermis or costa) will only be the child of the appropriate term.
If someone puts an annotation on leaf epidermis for rice, it will be collected by leaf, but not by vascular leaf, even though it should also be on vascular leaf.
Solutions:
- We will suggest to users that in cases like this, they annotate to both leaf epidermis and vascular leaf.
- Also, we will need to write an algorithm or script that will automatically add the annotations to vascular leaf, in case the curators miss it.
- It should search for annotations for parts of leaf, then check the taxon ID, and then add the appropriate annotation to either vascular leaf or non-vascular leaf.
- This will assure that annotations end up on the correct terms, but reduce term inflation.
It is likely that the same issue will come up for other classes, as we add terms and annotations for more taxa.
Treating different relations differently when dealing with annotations
Original request: March 9, 2011, but has been discussed earlier.
Under the current rules, all relations are treated equally, so all annotations get passed up from any child to its parent, regardless of the relation.
This works fine for is_a and part_of, but is problem for other relations.
develops_from: Annotations should not pass through the develops from relation. For example, leaf develops_from leaf meristem. If a gene is expressed in a leaf, this does not mean it is expressed in the leaf meristem that it developed from.
has_part: Annotations should pass through has_part backwards, that is from parent to child (in the opposite direction of the arrow). Furthermore, they should only pass through has_part relations when the instance for which the annotation was created comes from a taxon in which the parent term is part_of the child term.
For example: In anther has_part pollen sac, anther is the child, and pollen sac is the parent. Annotation should never go from anther to pollen sac. Annotations should go from pollen sac to anther, but only if they come from an angiosperms.
In order for this to work, the conditions under which annotations should pass through will have to be specified every time a has_part relation is added to the ontology. That means that any class in the PO that has a has_part tag will have to have additional tags specifying the conditions under which annotation should pass through the has_part relation. We would have to create a custom tag for this, perhaps a custom relation, that is conditional. Something along the lines of: if taxon_id=xxxx then treat parent term as part_of child term.
Linking images to PO
Original request: March 9, 2011, but this has been discussed on and off since 2010, including at the POC meeting at the NYBG in Nov. 2010.
Need to register Plantsystematics.org in the GO dbxref database so we can create shortcut links to their pages.
Problem with OWL File Generation
-OBO to OWL file generater is creating OWL file without the term definitions -Add further details etc
Register OBO_SF_PO in Go.xrf_abbs
We have a number of xrefs to OBO_SF_PO, which refer to the SourceForge tracker items associated with terms.